- Home
- About ACA
- Publications & Resources
- Programs
- Annual Meeting
- Membership
- ACA History Center
- Media Archive
|
By: David Rose Toy kits come in attractive boxes with the assembled product pictured on the outside. Inside are the small pieces, sometimes numbering in the hundreds, and detailed instructions on how to assemble them and in what order. Imagine if you didn’t know what those pieces were, how to put them together, or even what the final product looked like. That’s largely where we were just weeks ago with the SARS-CoV-2 coronavirus that causes COVID-19 disease. Already we know most of the pieces that make up the virus, some in great detail, how they are assembled into the whole, how the infection process works, and have ideas of how we might disrupt that process. How did we get to this point so quickly? Much of this results from Structural Science (Biology, Chemistry, Physics, Computation), including experimental studies by techniques such as crystallography, light scattering, and electron microscopy that allow us to learn about how atoms are organized in molecules, coupled with computational analysis comparing the genes and molecules of the virus to other known examples. This is only possible because of the fundamental research that created the tools and body of knowledge for this work, the existence of world-wide open and rapid scientific communication, and well-trained scientists ready and able to tackle the problems. Our society, the American Crystallographic Association, is a group of about one thousand members who use structural science to learn how atoms are arranged in all types of things around us. Why are viruses so dangerous compared to other microorganisms? This is because of their makeup: the molecules that constitute them. In contrast, bacteria, are living cells, with the molecules required for life encased within a cell wall, like a fortified, largely self-sufficient city. Conceptually, we have known how to combat bacteria for many years, often through molecules (antibiotics) that can break down those fortification walls. Other antibiotics attack the bacteria’s mechanisms for reproduction, such as how bacteria make their required proteins. Bacteria evolve comparatively slowly because mutations can affect the bacterial life cycle, itself. Nevertheless, some bacteria are evolving resistance to common antibiotics, and this is an important health threat; however, we have fundamental strategies how to approach them. Viruses are basically little packages of nucleic acids, RNA or DNA. DNA is, of course, what makes up our genomes, used by people interested in tracing their family background. Cells make copies of sections of our DNA in a different chemical form called RNA and use that to make proteins. Unlike bacteria, however, viruses contain little else beyond RNA or DNA and cannot reproduce and spread on their own; they rely on the host (patient) to do the work of making more copies of themselves. Viruses do not have a cell wall, nor do they have the molecules required to be self-sufficient. Therefore, the bacterial antibiotic approach won’t work, because viruses don’t need their nucleic acids to make the required molecules for life, their DNA or RNA can mutate freely.
How have we progressed from finding an unknown virus in late December 2019 to where we are now? Already, Structural Biology has told us a lot about the proteins in COVID-19 and how they work. There are several in common with other viruses, mostly involved in maintaining the RNA and holding the virus package together. Structural scientists in the US, among others, have already produced details of how the RNA is correctly processed (‘proofread’) by the viral proteins, and are studying how to inhibit these molecules. Other proteins, the most promising for targets for medical treatments, are the Spike and the Proteases. The Spike proteins, as the name suggests, are what gives the virus its distinctive crown-like protrusions (corona is Latin for crown, hence ‘coronavirus’). They are the Velcro-patches that roam around looking for ways to attach to our cells. In COVID-19, it seems that their main target (‘receptor’) in people is a protein called ACE2, which is common on cells in our respiratory system, hence the main means of entry into our body, and the main symptoms. Structural studies in China, the US, Germany and elsewhere, by electron microscopy at low temperatures (Cryo-EM) and by crystallography have identified the precise way that this interaction takes place, and that is leading to the next phase of trying to find safe drug molecules that block that interaction.
In the longer term, we know from experience having an effective treatment is not enough. This virus or related variants will not disappear, even once the pandemic is under control. As is true for influenza, development of a vaccine is the best way to control future outbreaks. A vaccine works by training our immune systems to create antibodies that recognize a virus and shut it down before it can reproduce. Structural Science is already playing a key role here. The structure of the entire COVID-19 virus has been predicted by computer analysis. While not based on experimental data directly, being able to see what parts of the virus are on the surface shows us what proteins might be used in a vaccine so that our cells can make antibodies for a virus our bodies have never seen. Doubtless, an actual experimental structure of COVID-19 is on the horizon, which is needed to support and refine the computer results. Looking to the future, we have already seen that this current pandemic is not a one-off; we have had regular, periodic outbreaks from new coronaviruses and they will continue. Once a vaccine is available, that will allow some level of control, as we have with ‘flu. But constant mutation and variation make these ‘moving targets’ and require regular vaccine refinements. These viruses will not be eliminated any time soon. Is there a more general approach that might address the common features of viral infection and, therefore, not be as sensitive to the specific virus structure? There is an intriguing, counter-intuitive approach that also relies on knowledge of the structure of molecules. What do these viruses have in common? They all rely on the host processes to reproduce and spread (propagate). If we can attack one or more key host activities that all viruses require, we may be on to a more general antiviral approach. But hang on a second: these processes are also needed for our own health. This is where structure comes in. The idea is that the virus uses a high level of rapid activity of some of these host molecules to propagate. However, for us to stay healthy, we only need a lower level of activity in many cases. Is there a ‘sweet spot’ where we can slow the virus down enough for our natural immune defenses to fight it, but still maintain enough activity for our own health? One example might be the process of glycosylation. Many of the viral proteins, such as the Spike protein, are not just protein, but also contain carbohydrate parts, so are ‘glycoproteins’. Without these carbohydrate parts (‘glycans’), the glycoproteins are often not made properly or are unstable. We have a strong fundamental knowledge from crystallography and other structural analyses, of the protein enzymes that build these glycans, and many inhibitors exist. Among the many molecules currently being tested as anti-virals are some of these glycosylation inhibitors. Whether this concept of host-targeted therapy is valid will require more research, including structural studies to improve the effectiveness of any promising compounds. But if it slows down the cycle pf pandemics or permits a broader anti-virus activity, it will be worth it, even once the current crisis subsides. Lessons so far:
A productive community of Structural Scientists depends on communication among researchers of the latest technologies and results. Since 1950, the American Crystallographic Association (ACA) has been the primary professional society for Structural Scientists in North America. The ACA depends for its existence on participation of the international scientific community and on donations.
Further information on the ACA, and to donate: https://www.amercrystalassn.org/ Scientific Links:Centers for Disease Control and Prevention: https://www.cdc.gov/media/subtopic/images.htm Spike protein: https://science.sciencemag.org/content/367/6483/1260 Main protease crystallography: https://science.sciencemag.org/content/early/2020/03/19/science.abb3405 Overview of COVID-19: https://www.sciencedirect.com/science/article/pii/S2090123220300540 Intact virus simulation: https://www.drugtargetreview.com/news/57287/3d-visualisation-of-covid-19-surface-released-for-researchers/ List of the structures so far: https://www.ebi.ac.uk/ena/pathogens/covid-19 |


 So, what do we know about the virus causing COVID-19 (SARS-CoV2)? Fortunately, from decades of fundamental research, we already knew a lot about the structures of viruses in general, and RNA viruses specifically, so that helped a lot. What does this beast look like and how does it infect us? The COVID-19 virus keeps a blueprint copy of itself as RNA. When we are infected, a RNA virus hijacks our cells to make more of its RNA, but that is not enough. RNA is a fragile type of molecule, quickly degraded even by exposure to air, so the virus also makes proteins to protect the RNA and to help infect new cells. These protein bits are the key to shutting down COVID-19.
So, what do we know about the virus causing COVID-19 (SARS-CoV2)? Fortunately, from decades of fundamental research, we already knew a lot about the structures of viruses in general, and RNA viruses specifically, so that helped a lot. What does this beast look like and how does it infect us? The COVID-19 virus keeps a blueprint copy of itself as RNA. When we are infected, a RNA virus hijacks our cells to make more of its RNA, but that is not enough. RNA is a fragile type of molecule, quickly degraded even by exposure to air, so the virus also makes proteins to protect the RNA and to help infect new cells. These protein bits are the key to shutting down COVID-19.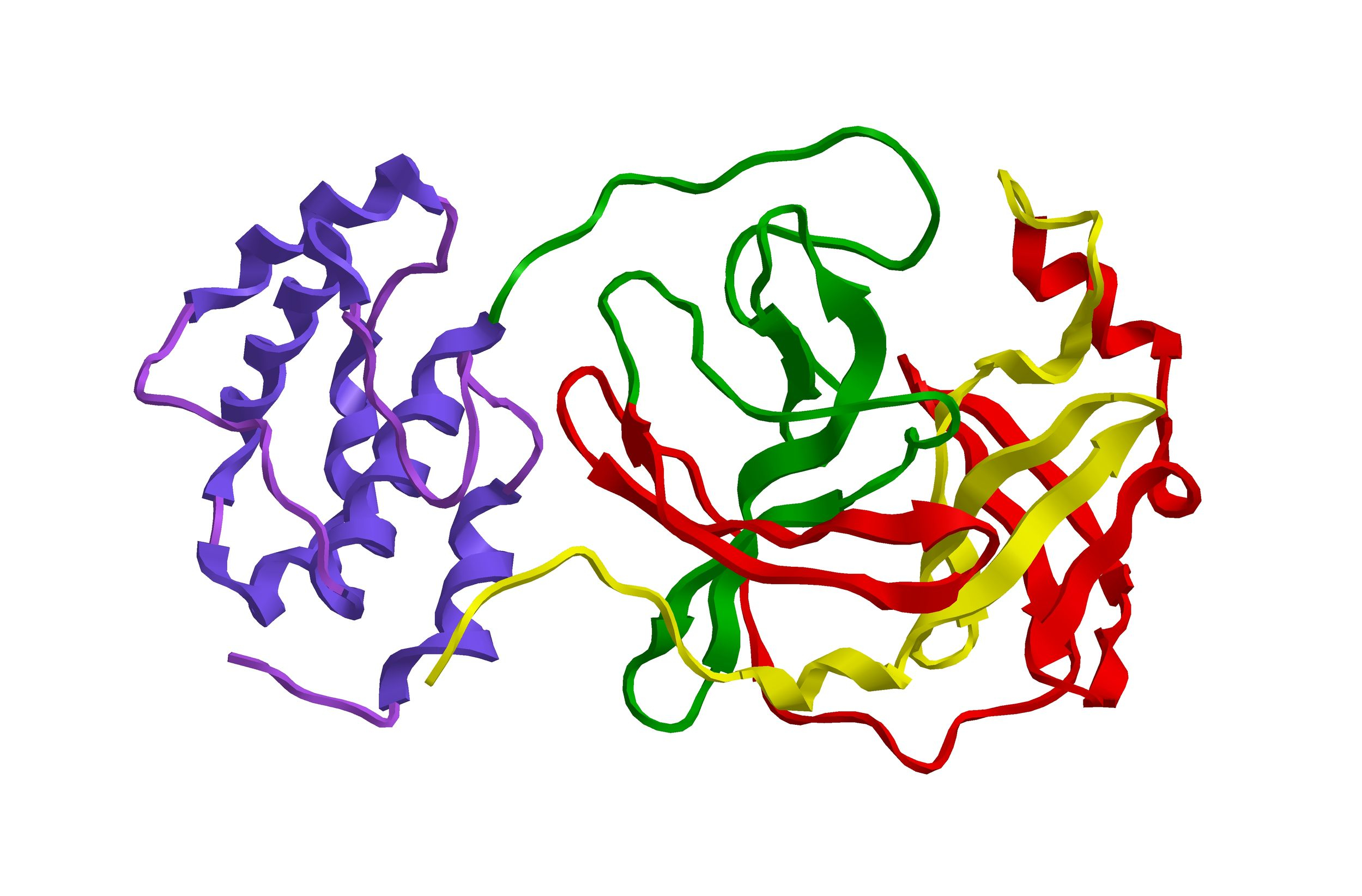 The proteins called Proteases (we now know there are two main proteases in COVID-19) have the job of taking the proteins that are made in the host cells under instruction from the virus RNA, and breaking those down into the individual proteins needed to make the new copies of the virus. Those are then packaged (again by hijacking our cells) into new virus particles, which can spread within the host and to infect other people. Proteases have a long history as targets for drugs in a family called therapeutic molecular inhibitors, so the COVID proteases have been synthesized in the laboratory and with record speed have been studied in great detail by structural methods like crystallography. The virus needs both proteases, so inactivating one seems to be enough to stop the infection; however, structures of both are already available. Several research groups around the world have already screened libraries of molecules where most have been tested as possible treatments for other diseases, so they are already known to be safe in humans, and found some ‘hits’ that seem to be effective at inhibiting one or other of the Proteases. The complexes formed when a drug binds to and shuts down the proteases have also been examined by crystallography, propelling chemists to design improvements in activity and efficient means of synthesis. The role of the Crystallographers and Structural Scientists in rapidly mobilizing their efforts, and openly reporting their results, cannot be overstated.
The proteins called Proteases (we now know there are two main proteases in COVID-19) have the job of taking the proteins that are made in the host cells under instruction from the virus RNA, and breaking those down into the individual proteins needed to make the new copies of the virus. Those are then packaged (again by hijacking our cells) into new virus particles, which can spread within the host and to infect other people. Proteases have a long history as targets for drugs in a family called therapeutic molecular inhibitors, so the COVID proteases have been synthesized in the laboratory and with record speed have been studied in great detail by structural methods like crystallography. The virus needs both proteases, so inactivating one seems to be enough to stop the infection; however, structures of both are already available. Several research groups around the world have already screened libraries of molecules where most have been tested as possible treatments for other diseases, so they are already known to be safe in humans, and found some ‘hits’ that seem to be effective at inhibiting one or other of the Proteases. The complexes formed when a drug binds to and shuts down the proteases have also been examined by crystallography, propelling chemists to design improvements in activity and efficient means of synthesis. The role of the Crystallographers and Structural Scientists in rapidly mobilizing their efforts, and openly reporting their results, cannot be overstated.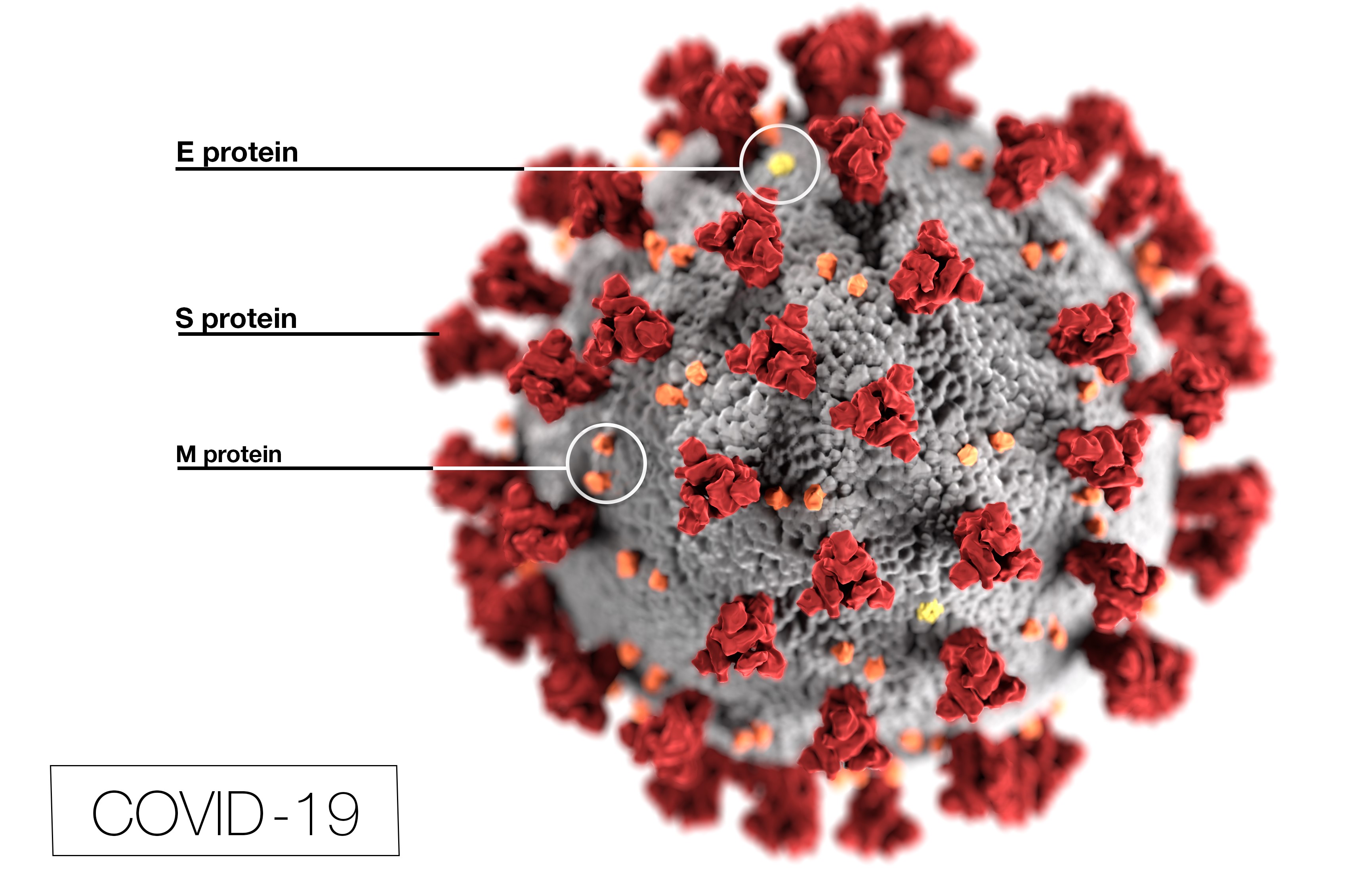 This visualization of the outside shell of the COVID-19 virus, from the Centers for Disease Control and Prevention, shows the Spike protein (S) on the virus surface in red, and other exposed proteins (M and E) that could be accessible as vaccine targets. The RNA is protected inside the shell.
This visualization of the outside shell of the COVID-19 virus, from the Centers for Disease Control and Prevention, shows the Spike protein (S) on the virus surface in red, and other exposed proteins (M and E) that could be accessible as vaccine targets. The RNA is protected inside the shell.


















