- Home
- About ACA
- Publications & Resources
- Programs
- Annual Meeting
- Membership
- ACA History Center
- Media Archive
ACA Early Career Scientist Spotlight-23
Personal StatementMy research is centered on proteins involved in DNA alkylation damage repair. DNA damage occurs thousands of times per cell per day, including alkylation damage that is induced by several factors such as UV radiation and chemicals that we encounter daily. Studying this I have used site directed mutagenesis and NMR spectroscopy to predict and test for residues that are at the binding interface between ASCC2 and K63-linked diubiquitin, as well as how the specificity between the two is achieved. Based on previous NMR experiments I predicted residues that are likely at the binding interface and mutated those residues on ASCC2 and the proximal ubiquitin. The contents of my poster were work conducted by several members of the lab, current and alumni, and with advice from Dr. Lombardi. The ITC experiments were conducted in Dr. Cynthia Wolberger’s laboratory, and the 3D NMR experiments were conducted by Dr. Ananya I joined the Lombardi lab as a rising sphomore in 2020. Although COVID-19 reduced the time spent in the lab, I was still able to experience in-person benchwork and begin the journey of my development in research, which I’ve grown to appreciate more due to radical changes that occurred at the onset of the pandemic. The lab now consists of Dr. Patrick Lombardi and ~15 other undergraduates, myself included. This field of research is exciting to me because there are different things that can be pursued in the same pathway using several different experimental techniques which I have continued to learn over time. Outside of my courses and lab, I enjoy working as a supplemental instructor, reading supernatural fiction books, watching paranormal TV shows and chick-flicks, and organizing and attending events for the French and Culture club at the Mount. I also enjoy my French courses very much and love to read outside on a swing. My favorite places on campus are the lab, and the outdoor seating outside of the Coad science building. When explaining my research to family and non-science friends, I usually say I look at a pathway that repairs a certain type of damage in DNA. And to do this, I make small changes to some of the proteins and test for changes in the behavior of the proteins. This allows me to see if that particular part of the protein is important for how the pieces of the pathway come together. What do you think? |


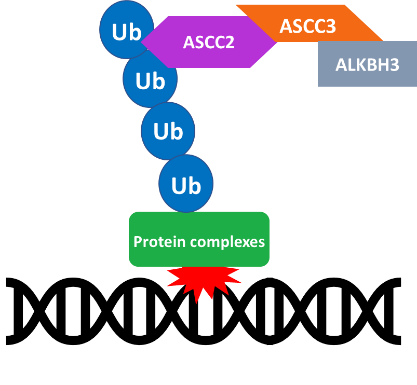 pathway is important as it can inform certain treatments like alkylation therapy. There are several repair pathways involved in alkylation damage. In the Lombardi lab, we study one such repair pathway that includes the ALKBH-ASCC repair complex. This pathway consists of polyubiquitin chains that are assembled at damage sites and the complex which includes multiple ASCC components (1, 2 and 3) and ALKBH3. We work to determine the interaction between the binding component of the subunit (the ASCC2 CUE domain) and the K63-linked polyubiquitin chains.
pathway is important as it can inform certain treatments like alkylation therapy. There are several repair pathways involved in alkylation damage. In the Lombardi lab, we study one such repair pathway that includes the ALKBH-ASCC repair complex. This pathway consists of polyubiquitin chains that are assembled at damage sites and the complex which includes multiple ASCC components (1, 2 and 3) and ALKBH3. We work to determine the interaction between the binding component of the subunit (the ASCC2 CUE domain) and the K63-linked polyubiquitin chains.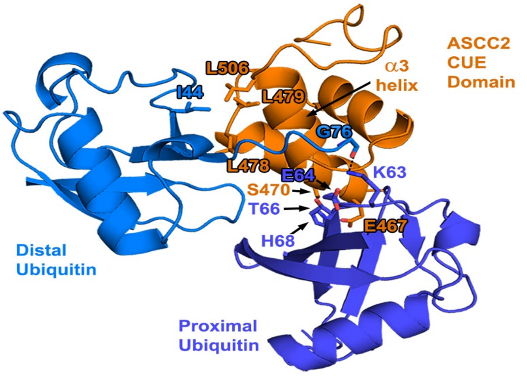 Majumdar, both at the Johns Hopkins School of Medicine in Baltimore, MD. The protein labeling for 3D NMR was done my colleagues Zachary Beck, Angelo Gurcsik, Devin Shorb, and me. It was a great pleasure to present our work at the 2022 ACA meeting.
Majumdar, both at the Johns Hopkins School of Medicine in Baltimore, MD. The protein labeling for 3D NMR was done my colleagues Zachary Beck, Angelo Gurcsik, Devin Shorb, and me. It was a great pleasure to present our work at the 2022 ACA meeting.


















