ACA Early Career Scientist Spotlight-24
 |
Shi Feng
Cornell University
Graduate Student
ACA Member Since 2023
Shi Feng is biophysics graduate candidate co-advised by Dr. Richard Cerione and Dr. Brian Crane in the Department of Chemistry & Chemical Biology at Cornell University. Originally from Xuancheng, China, he obtained his B. Eng. in Biomedical Engineering in 2018 at Southeast University, Nanjing, China. His research involves the application of cross-disciplinary methods to study the molecular mechanisms that regulate cancer biology and circadian clocks.
|
Personal Statement
Since I was a kid, light always fascinates me with its unpredictable colors but untouchable reflection, which leads my efforts to not only visualize the protein using structural biology, but also the molecular mechanism of photo receptor. One of my projects with Dr. Brian Crane is to understand how photo receptor regulate the timing mechanism of circadian rhythms.

The circadian clock of Drosophila melanogaster has served as the paradigm for understanding the basis of circadian rhythms of higher organisms, as recognized by the 2017 Nobel Prize in Physiology and Medicine to Hall, Rosbash and Young. Using this model system, I discovered the structural basis for how light-sensing Cryptochrome (Cry) recognizes the clock corepressor/cotransporter element Timeless (Tim). The structure of the Cry-Tim complex that I determined revealed key aspects of how Cry engages its targets to set the clock, and how Tim gates nuclear entry to regulate the clock period, which are both important advancements in the fields of circadian clocks and photosensory transduction. Cry is a highly conserved signal transduction protein, closely related to blue-light sensing photolyases, also flavin-binding proteins that repair DNA damage. By determining the 3D structure of the Cry-Tim complex, we found that the N-terminus of Tim inserts into the flavin binding pocket of light-activated Cry, mimicking how 6-4 photolyase binds to damaged DNA. This binding mode also helps to explain the temperature and latitude adaptation of the circadian clock of flies. We also found a secondary binding site in the disordered region of Tim, which could potentially provide a rationale for certain sleep disorders that occurs in humans. Finally, a newly discovered region in the Tim groove suggests the mechanism by which Tim is imported into the nucleus to suppress the translational-transcriptional feedback loop and thereby provide key delays for setting the clock period. This work is recently published in Nature.
 |
|
Lin, C.*, Feng, S.*, DeOliveira, C. C., & Crane, B. R. (2023). Cryptochrome–Timeless structure reveals circadian clock timing mechanisms. Nature, 617(7959), 194-199. (*co-first authors)
Cryo-EM structure of the Cry–Tim complex. (a). Drosophila Cry entrains the circadian clock by initiating degradation of Tim in light. Jet, Per, Doubletime (Db), Cyc, Clk and Impα1 compose the core circadian oscillator. hν represents photon energy, in which h is the Planck constant and ν is the photon frequency. (b). Cry–Tim docked into the electron density map.
|
Besides photo receptor, I am also generally interested in the protein assembly dynamics in the cancer signaling transduction and metabolism. I am studying the activation mechanism of a key enzyme in cancer cell metabolism, glutaminase, which catalyzes the hydrolysis of glutamine to glutamate and thereby provides cancer cells with an essential source of carbon and nitrogen. Thus, targeting glutaminase is a promising approach to develop new therapies against several different types of cancer. Using structural biology as well as complementary biophysics and biochemical tools, I have made the exciting discovery that glutaminase activity is coupled to its ability to from filaments of repeating tetrameric units. These findings provide us with new insights into the mechanism that promotes the catalytic activity of glutaminase. Moreover, they identified a new druggable pocket that could potentially be disrupted to block glutaminase filament formation and activity, which will be a focus of my future work. This work is recently accepted by Nature Communications.
Within my remaining time in graduate school, I will continue working on the photoreceptor and signal transducing proteins. I would like to thank my supervisors Dr. Richard Cerione and Dr. Brian Crane for their great mentorship, and my colleagues Cody, Tien, Shawn, Changfan and Christina for their help in the projects.
 |
|
Feng, S., Aplin, C., Nguyen, T. T. T., Milano, S. K., & Cerione, R. A. (2023). Filament formation drives catalysis by glutaminase enzymes important in cancer progression. bioRxiv, 2023-02. (Accepted by Nature Communications)
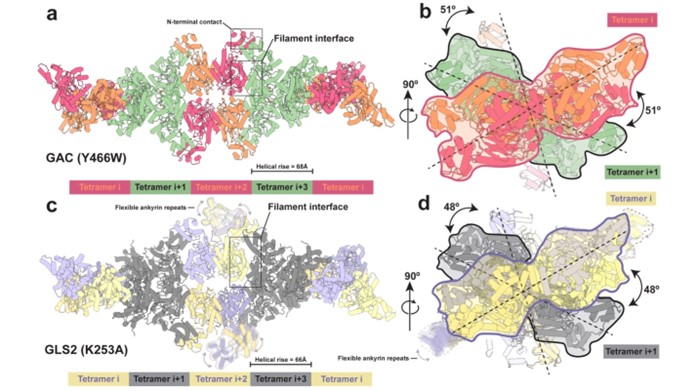
Structures of the human GAC (Y466W) filament and the human GLS2 (K253A) filament captured under catalytic turnover conditions.
(a) Structure of the GAC (Y466W) filament showing a five tetramer stretch of the higher-order oligomer with a helical rise of 68 Å. Tetramers are colored alternately either orange and red or green. The orange and red tetramers are colored to distinguish between monomers in each tetramer. The filament interface and the N-terminal contacts are highlighted. (b) The GAC (Y466W) filament viewed down the filament axis. The first two tetramers are outlined with red and black silhouettes, sequentially. The dashed line indicates the longest dimension of each tetramer and the 51° rotation around the filament axis between tetramers. (c) Structure of the GLS2 (K253A) filament showing a five tetramer stretch of the higher-order oligomer with a helical rise of 66 Å. Tetramers are colored alternately either purple and yellow or gray. The purple and yellow tetramers are colored to distinguish between monomers in each tetramer. Ankyrin repeats are not built due to their high flexibility and outlined with dashed profile. The filament interface is highlighted. (d) The GLS2 (K253A) filament viewed down the filament axis. The first two tetramers are outlined with purple and black silhouettes, sequentially. The dashed line indicates the longest dimension of each tetramer and the 48° rotation around the filament axis between tetramers.
|
|
























