ACA Young Scientist Spotlight-21
Rohit Jain | Postdoctoral Scholar | Case Western Reserve University | ACA Member Since 2020
Rohit's research focus is on understanding protein complexes in their physiological environment, an essential aspect of cell survival. In this quest, he has investigated protein structure at multiple levels, from amino acids to peptides to proteins to protein complexes. Rohit hasexpertise in several cutting-edge biophysical techniques like Mass spectrometry, Protein footprinting and X-ray scattering. His doctoral thesis was at Max Planck Institute for Biophysical Chemistry. Here, he utilized high flux X-ray synchrotrons to decipher structural dynamics of proteins, ranging from structural freezing to milliseconds to <100 microseconds timescales. In his time as a postdoc at UMass Medical School, Rohit investigated the role of amino acid sequence in modulating the folding free energy landscape of TIM barrel motif, which is one of the most ancient and common motifs in biology. Presently, Rohit is utilizing protein footprinting to elucidate molecular structure and binding behaviors of biological recognition elements with stress biomarkers and neuropeptides.
Three Fun Facts About Young Scientist Rohit Jain
-
Walking on beach is his stressbuster.
-
His name means orange sunlight during early morning and sunset.
-
Loves Ice Cream
Personal Statement
Most of the work presented in mine ACA 2021 poster was done at a small corner in my home due to ongoing COVID-19 pandemic. It has been difficult to continue to stay motivated during this time and acknowledgment of my research during ACA 2021 conference has fueled my ambition to do world class science and help human society. I feel that last year, which has been tragic worldwide, has brought the usefulness of basic and applied science to the front page of every news and the living room of each family. My hope is that the ongoing question about importance of doing science will now subside and we will be able to resolve biomedical challenges facing human society.
My research is based on curiosity about how macromolecules work in their cellular environment. Seminal discoveries like C. B. Anfinsen’s work on folding of protein chain and R. D. Kornberg’s work on understanding the molecular basis of eukaryotic transcription have inspired me to work on protein structure-function relationship. I have utilized X-ray based techniques namely X-ray footprinting, X-ray crystallography and X-ray scattering to understand protein folding and structure for biomedically important targets. Presently, I am working towards biosensor development for real time monitoring of stress in unmet human health needs. For reaching this goal, I and my colleagues have elucidated lactate oxidase enzyme-biomarker lactate interactions by small-angle X-ray scattering, X-ray crystallography and X-ray footprinting (Reference ACA 2021 poster). The X-ray footprinting of lactate oxidase utilized a high throughput endstation at XFP beamline in NSLS II (NY, USA), where, I helped develop a robust, easy-to-use platform for carrying out X-ray footprinting experiments (Figure 1-below).
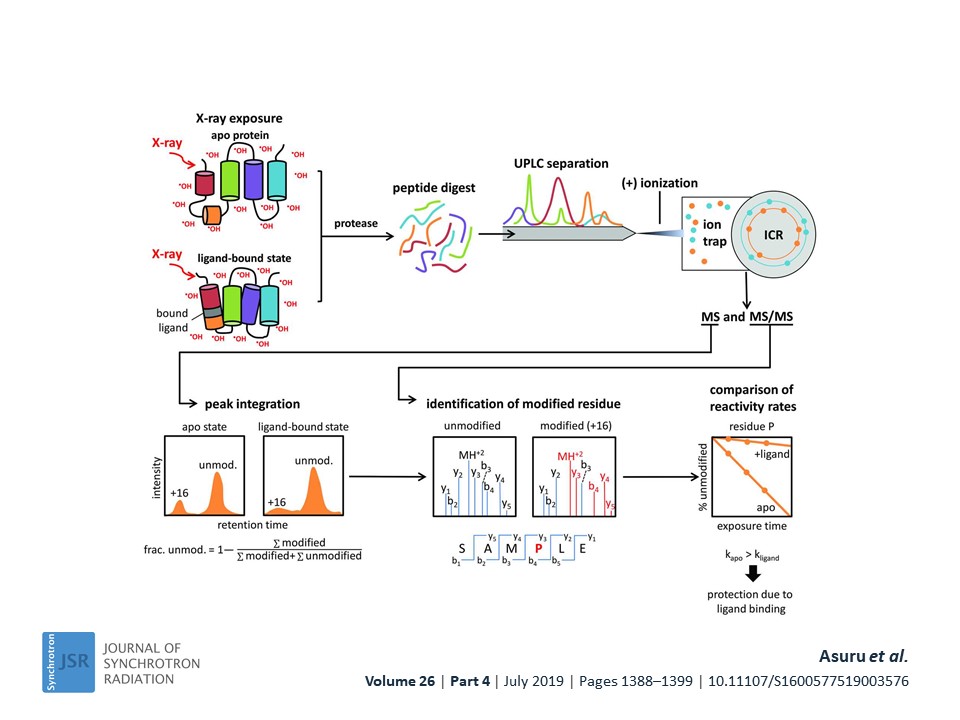
Using this high throughput endstation, I illustrated the hydroxyl scavenging properties of 26 compounds commonly encountered in biochemical sample preparation (PMID: 34475281). This development has increased efficiency for structural biology research with X-ray footprinting by allowing exposure of thousands of samples in a day at the premier synchrotron facility NSLS II. In one of my most gratifying research project, I found the evolutionary conservation of protein folding mechanisms and structures of partially folded states for orthologous IGPS TIM barrels (PMID: 33875592). First, I mapped the 6-state folding free energy surface of a ~3.6 billon year old, bacterial IGPS TIM barrel by hydrogen exchange mass spectrometry and complementary biophysical techniques. Then, I performed bioinformatics analysis of thousands of IGPS sequences from three superkingdoms of life to reveal an exceedingly high hydrophobicity and surprisingly alpha-helix propensity for Beta4 strand (Figure 2-below).
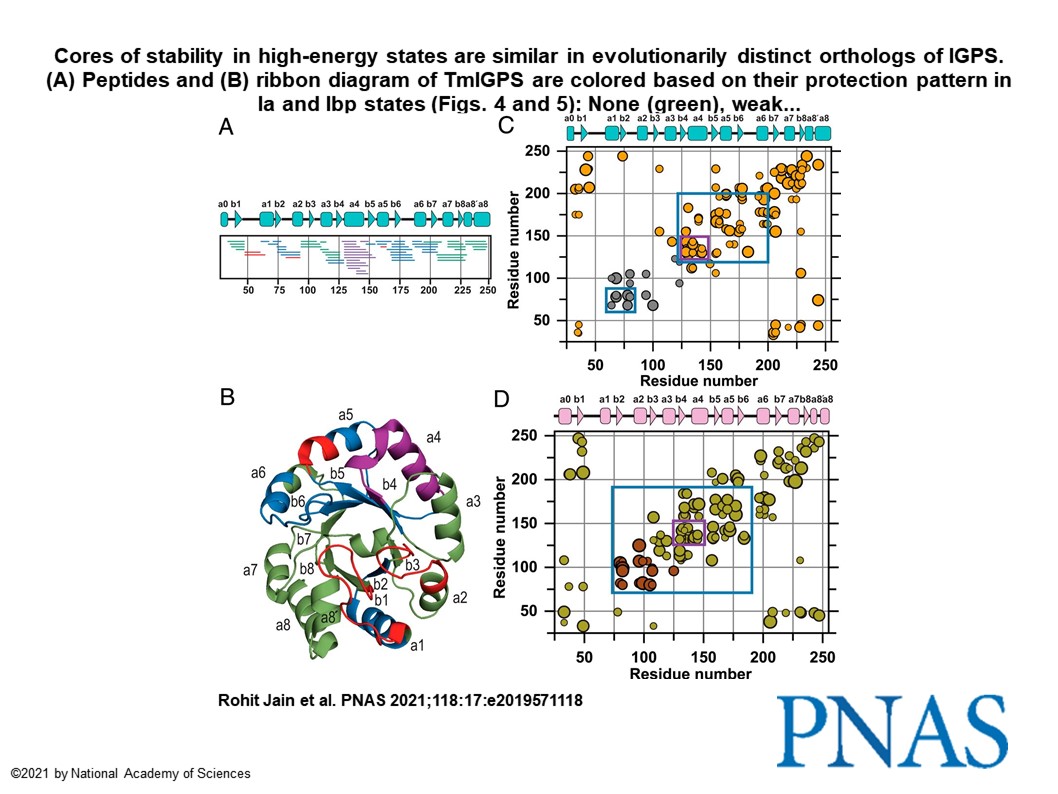
This asymmetry in the highly favored aliphatic sequences for (βα)4 segment in IGPS TIM barrels with eight βα segments could be useful for the de novo design of TIM barrels, a quest for >25 years. This development will allow computer coded proteins to achieve efficient and rapid folding while avoiding aggregation.
My present focus on X-ray footprinting is a full circle from my PhD thesis on insights into protein dynamics by X-ray scattering. During PhD at world renowned Max Planck Institute of Biophysical Chemistry, I utilized high-flux photons from advanced X-ray synchrotrons to decipher the structural dynamics of biomedically important proteins, ranging from structural freezing to milliseconds to <100 microseconds timescales. My work on structural freezing of human Guanylate kinase was the first to uncover structural details (Figure 3-below) for this important anticancer and antiviral therapeutic target (PMID: 26446352).
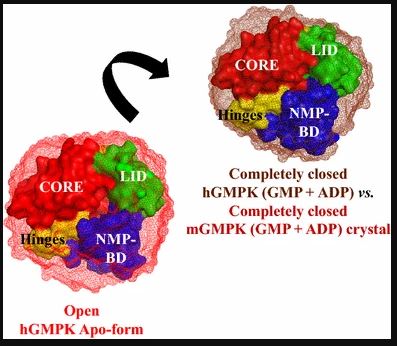
I combined a microfluidics based fast micromixing device and micrometer focused X-rays at cSAXS beamline (PSI, Switzerland) to probe the millisecond unfolding of a protein folding model “ubiquitin” with much reduced sample consumption (PMID: 24092048). Utilizing this setup, I obtained structural-kinetics-thermodynamics insights into ubiquitin unfolding reaction and they helped to understand the origin of non-linear kinetics during ubiquitin unfolding. My research on time-resolved X-ray scattering will help to resolve the structural dynamics of proteins on nanometer scale and on timescales extending over many orders of magnitude down to the femtosecond range (PMID: 26732244).
The excitement of discovery of unknown and pure joy of troubleshooting in research projects have motivated me to work on challenging research projects. I have found presenting and discussing my work during conferences to be both interesting and helpful. Out of all presentations, explaining my research to non-scientists has been most eye opening for me. For example, explaining what proteins are and how they function, I often have used examples of machines from daily lives. This has helped me noticed greater subtleties and significance of my research.
|