- Home
- About ACA
- Publications & Resources
- Programs
- Annual Meeting
- Membership
- -History-
- ACA Video Library
ACA Early Career Scientist Spotlight-24
Personal StatementAlthough I was born in the United States, I spent my youth in the countries of the Dominican Republic and Panama, making Spanish my first language. I remember coming back to the U.S. and struggling in some courses due to the language barrier. However, I found myself succeeding in mathematics and in the sciences. Mathematics, biology, chemistry: they had the same numbers and truths in both languages. I found myself having a high affinity to these subjects both because I could understand them even with my gap in English and because I felt things just “clicked.” I still now often say the first language I learned was math, not English. This affinity towards STEM continued throughout all of my education, and I now find myself pursuing a Ph.D. in chemistry in the Smith Lab at the University of Maryland, Baltimore County (UMBC). Each day teaches me something new, and I become more intrigued by science as each day goes by.
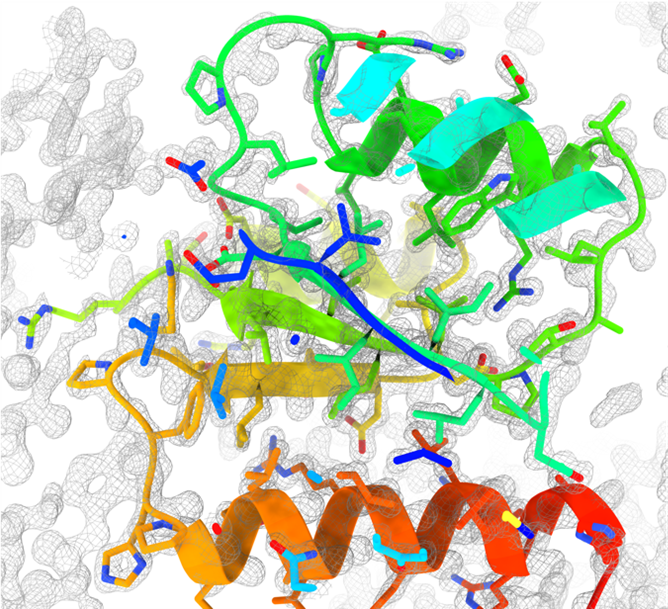
In the Smith lab at UMBC, I research bioinorganic chemistry, which is a field of science that studies the roles of metals in biology. More specifically, I am currently working on the structural and biophysical characterization of a two-component signal transduction system that regulates biofilm formation/decay in Pseudomonas aeruginosa through the binding and the sensing of extracytoplasmic Fe2+. Biofilms are complex microbial structures capable of providing an advantageous protective quality that allows bacteria living within a biofilm to be significantly more resistant to antibiotic treatments than planktonic bacteria, representing a major health threat to the world. The two proteins comprising the two-component system I study work in tandem to regulate the transcription of multiple virulence factors, and a greater understanding of these proteins could allow for novel therapeutics to be made to battle antimicrobial resistance. I have spent the majority of my dissertation research biophysically and biochemically characterizing this metal-sensing two-component system, that comprises a membrane-bound histidine kinase (HK) and a soluble response regulator (RR). For the membrane HK, I have worked to understand the mechanism of metal sensing of this complex membrane protein. For soluble RR, I have crystallized and solved the N-terminal effector domain of this protein to 1.3 Å resolution, which represents the first structure I have ever solved.
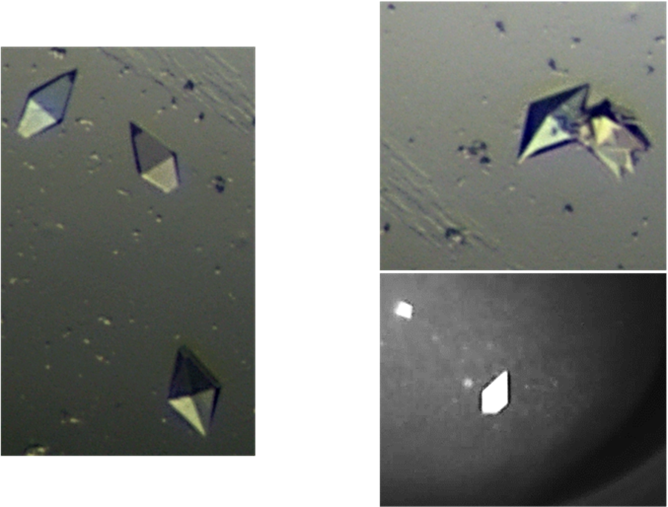
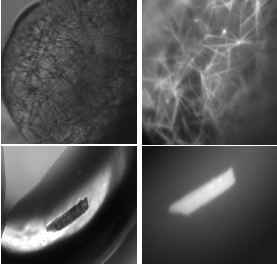
What interests and excites me the most about crystallography is how it is both a science and an art. My labmates and I always say that crystallography needs some skill, plenty of patience, and plenty of luck. Mixing specific components of a precipitant and making slight variations until you have come up with the best crystallography condition is where all the art comes to life. It is almost like every crystal is making a new Powerpuff Girl, composed of a little sugar, spice, and everything nice (to crystallize!). This is also how I often explain crystallography to non-crystallographers: you combine just the right “special spices” to make the perfect crystal. Since solving the soluble RR structure, I have been working on crystallizing other proteins, testing the crystals for diffraction, and trying to solve additional structures. If crystallography teaches you anything its persistence!
I am also not the only one working through structural endeavors. While the Smith lab does not consider itself a “purely structural” lab, every one of my labmates is working on structurally characterizing their own biological macromolecule, whether that be through X-ray crystallography, NMR, cryo-EM, or SAXS. We are all studying the structure-function relationships of various metalloproteins, and we especially focus on iron metalloproteins. My research lab is made up of a fantastic group of bioinorganic chemists and biochemistry, and we are constantly bouncing structural ideas off of each other for improving construct design, crystal looping, cryo freezing, diffraction strategies, and more. Our overall goal is to leverage our understandings of these critical metalloproteins to treat human diseases and to improve human health. 
All things considered, I would definitely recommend crystallography to any researcher. Crystallography is hard work but it is extremely rewarding. My labmates and I often spend months tweaking one condition to improve resolution, spend weekends and evenings having late-night beam trips, and we even spend days if not weeks refining one structure. However, solving that structure in the end always makes it worth it! |
|||||||